Modeling Protein Structure
Harry S. Truman High School, Bronx
Summer Research Program for Science Teachers
August 2010
Subject:
Biology
Aim: How do the properties of amino acids affect the shape of proteins?
Objectives: SWBAT
- Explain that amino acids are the building blocks of proteins
- Understand that there are 20 different amino acids
- Discuss the properties of polar (hydrophilic, negatively charged/acidic or positively charged/basic) and non-polar (hydrophobic, uncharged) side chains (R-groups) of these amino acids have important properties
- Demonstrate the secondary folding (alpha helices and beta pleated sheets) and tertiary folding of peptides
- Demonstrate that protein tertiary structure is created when the secondary structures fold and form bonds to stabilize the structure into a unique shape
Materials: Mini-Toobers, handout with molecular structure of 20 amino acids, molecular attachments for the Mini-Toober (or colored thumbtacks), scissors
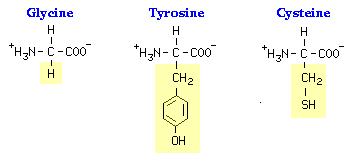
Do Now: Give each student a handout with 6 amino acids. Have students discuss which amino acid R-groups could react with each other by forming bonds. What kinds of bonds? How could these bonds affect the shape of a polypeptide chain?
Lesson Overview: Students
work in small groups. Each group
will be given a Mini-Toober (available from
3DMolecularDesigns.com) and several
attachments representing R-groups of amino acids (e.g. hydrogen molecules, SH
groups, polar side chains and non-polar side chains). (Colored
thumbtacks can also be substituted to represent these molecules.)
Ask students to
fold half the Mini-Toober to create a beta-pleated sheet and the other half to
create an alpha helix. Elicit from
the class that these secondary structures are created through hydrogen bonds.
Discuss how amino acid sequence effects the structure and
function of proteins.
Optional:
Following this activity, give each group a specific sequence of 6 amino acids to
represent on their Mini-Toober. (These amino acids can be on a Xerox handout and
students can cut them out with a scissors.)
Students must add the appropriate R side chains to their Mini-Toober and
then try to fold it to create bonds that make sense
(e.g. 2 cysteine molecules would form a disulfide bridge, adjacent
glycine and tyrosine R groups may form a hydrogen bond).
“Proteins
are polymers of amino acids covalently linked through peptide bonds into a
chain. Within and outside of cells, proteins serve a
myriad of functions, including structural roles (cytoskeleton), as catalysts
(enzymes), transporter to ferry ions and molecules across membranes, and
hormones to name just a few.
“With few exceptions, biotechnology is about understanding, modifying and ultimately exploiting proteins for new and useful purposes. To accomplish these goals, one would like to have a firm grasp of protein structure and how structure relates to function. This goal is, of course, much easier to articulate than to realize! The objective of this brief review is to summarize only the fundamental concepts of protein structure.
Amino Acids

“Except for glycine, which has a hydrogen as its R-group, there is asymmetry about the alpha carbon in all amino acids. Because of this, all amino acids except glycine can exist in either of two mirror-image forms. The two forms – called stereoisomers – are referred to as D and L amino acids. With rare exceptions, all of the amino acids in proteins are L amino acids.
“The unique side chains confer unique chemical properties on amino acids, and dictate how each amino acid interacts with the others in a protein. Amino acids can thus be classified as being hydrophobic versus hydrophilic, and uncharged versus positively-charged versus negatively-charged. Ultimately, the three dimensional conformation of a protein - and its activity - is determined by complex interactions among side chains. Some aspects of protein structure can be deduced by examining the properties of clusters of amino acids. For example, a computer program that plots the hydrophobicity profile is often used to predict membrane-spanning regions of a protein or regions that are likely to be immunogenic.
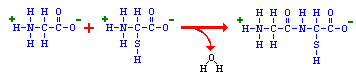
“The head-to-tail arrangement of amino acids in a protein means that there is a amino group on one end (called the amino-terminus or N-terminus) and a carboxyl group on the other end (carboxyl-terminus or C-terminus). The carboxyl-terminal amino acid corresponds to the last one added to the chain during translation of the messenger RNA.
Levels of Protein Structure
“Structural features of proteins are usually described at four levels of
complexity:
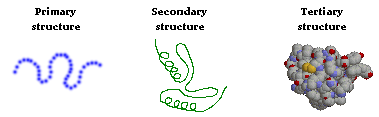
“The primary structure of a protein can readily be deduced from the
nucleotide sequence of the corresponding messenger RNA. Based on primary
structure, many features of secondary structure can be predicted with the aid of
computer programs. However, predicting protein tertiary structure remains a very
tough problem, although some progress has been made in this important area.”
New York
City Performance Standards:
S1a, S1b, S1c, S2a, S2d, S4a, S5a, S5c, S6a, S7b, S7c