Using VNTR analysis to Identify Guilt in at a Crime Scene
Rachel M. Lytle
High School of Telecommunication Arts and Technology
Summer Research Program for Science Teachers
Summer 2006
Subject: AP Biology/ Forensics
Grade Level: 9th through 12th
Unit: Genetics, Biotechnology, lab skills
Objective: Students will be able to:
denaturation, primers, replication, template, polymerase, PCR, gel electrophoresis, VNTR, autoradiogram
Prior Knowledge: Teacher discussion on DNA replication, protein synthesis and the function and structure of the DNA molecule, gel electrophoresis, pipeting skills, solution preparation.
New York State Science Standards
Standard 1 Key Idea 1-performance indicator 1.1b
Key Idea 2-performance indicator 2.2a
Standard 4 Key Idea 2-performance indicators 2.1a, 2.1b, 2.1c, 2.2c, 2.2d, 2.2e
Key Idea 5- performance indicators 5.1f, 5.1g
National Science Learning Standards met by this lesson:
Background Information:
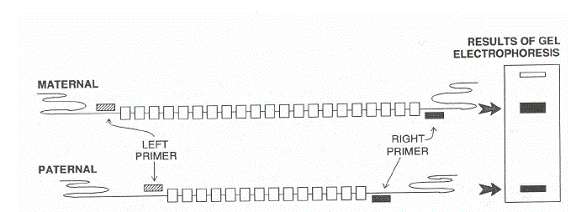
A tandem repeat is a short sequence of DNA that is repeated in a head-to-tail fashion at a specific chromosomal locus. Tandem repeats are interspersed throughout the human genome. Some sequences are found at only one site -- a single locus -- in the human genome. For many tandem repeats, the number of repeated units varies between individuals. Such loci are termed VNTRs.
In diagram below one person has inherited the allele for 21 copies of a repeated sequence from its mother and 15 copies of the same sequence from its father.

When profiles from a single VNTR locus from unrelated individuals are compared, the profiles are normally different. However it is possible for two individuals to have the same profile at one or two loci by chance. But the chance of more than one person having the same DNA profile at 4, 5, or 6 different VNTR loci is extremely small. When DNA
Lab part A
Day 1- Isolate your DNA from cheek cells,
Day 2- Set up PCR reaction
Day 3—Cast Agarose gel
Day 4- Run Gel Electrophoresis
Day 5- Photograph gel and analyze data
Day 6- Begin Probability Activity
Day 7- Complete Probability Activity
Day 8- Write recommendation to the Dean
Materials Needed
Thermocycler or 3 water baths
Centrifuge for 15mL tubes
Microcentrifuge
Hot Plates and 600 ml beakers
Adjustable micropipette
Electrophoresis chamber
Power supply
UV transilluminator
Camera to picture gel
Test tube racks
Supplies per class
Paper cup for each student
15 ml tube for each student
1.5 ml tube for each student
PCR tube for each student
Micropipets
Permanent lab markers
Reagents
300ml- 0.9% NACl solution
Chelex
MgCl2
Distilled water
Agarose
Electrophoresis buffer
Loading dye
Ethidium bromide
Teacher Preparation prior to lab
Student Procedure
Part 1- Isolating Your DNA from Cheek Cells
Part 2: Setting up your PCR- making copies of your DNA for analysis
|
Check when added |
||
|
|
Cheek DNA |
10uL |
|
|
PCR Mix |
40uL |
> This solution contains 1uM of each D1s80 primer, the 4 dNTP’s (A,T,G,C) and Taq polymerase
|
|
Temperature |
Time |
|
|
Soak |
94oC |
1 minute |
This step bring the samples to the correct temperature |
|
30 Cycles |
94oC |
15 seconds |
This temperature denatures the DNA |
|
|
68oC |
15 seconds |
Primers anneal to the template during this incubation |
|
|
72oC |
15 seconds |
New DNA is synthesize |
|
Termination |
72oC |
10 minutes |
|
|
|
4oC |
|
Low temperature ensure the stability of the PCR products |
Questions
The sequence of the D1S80 Primers is given below. Right the DNA sequence they will bond complimentary to.
Primer 1
5’-GAAACTGGCCTCCAAACACTGCCCGCCG-3’
Primer 2
5’- GTCTTGTTGGAGATGCCCCTTGC-3’
1. Explain why Taq polymerase is used to : "copy" human DNA in PCR and not human polymerase.
2. Describe 3 ways in which PCR is similar to DNA replication.
3. Describe 3 ways in which PCR is different than DNA replication.
Part 3: Cast 2% Agarose Gel- One gel will be made for every 8 students
|
Ethidium bromide is a mild carcinogen, so you should always wear gloves when working with it. The ethidium bromide will make our DNA visible under UV light. |
Answer the following questions about DNA gel electrophoresis
Part 4- Run your PCR amplified DNA samples using Gel Electrophoresis
Part 5-Visualize Gel
Results and Discussion
Briefly describe below what information you gained from examining your gel.
At 3:01PM on October 23 of this year Mr. Michael the school’s meanest math teacher was staying after school to grade tests. Many students did not like Mr. Michaels because he assigned tons of homework and gave his students the toughest test. You are two other students were sitting in the room completing a test you had missed. Mr. Michael’s wife called his cell phone so he stepped out into the hallway to use his phone. When Mr. Michael returned he found a large glob of spit floating in his coffee. Anxious to catch the culprit Mr. Michael took you, the two other students, and the coffee with spit in it to Mrs. Lytle’s science classroom. Mrs. Lytle agreed to help Mr. Michaels determine who spit in his coffee using VNTR analysis. The audioradiograms Mrs. Lytle made using all the DNA samples have been provided to you. Using the data provided calculate the probability at which you, and the two other students could present with such a DNA profile. Present your findings to the school dean so that he can take proper disciplinary action.
At this point we do not know whether this is very strong evidence that the suspect committed the crime. We need to know whether such a match is likely to have occurred simply by chance, or whether it is reasonable to conclude that the multilocus genotype matches because the evidence was in fact left by the suspect. So we will calculate the probability of such a five-locus genotype occurring by chance in the population.
We will use a process called “binning”. In human DNA, because there are so many different sized alleles, some of which are very similar in size, it would be impossible to try to distinguish every different allele size on a gel of this kind. To get around this problem, bands that fall between adjacent markers are classified as all belonging to the same “bin”, even if they differ somewhat in size.
You have been given tables of actual allele frequencies from a Caucasian reference population for the three different VNTR loci used in this case, presented on the same page with the autoradiogram showing the pattern observed at that locus. These tables show the alleles sizes that were observed when these loci were scored in large samples of individuals. You can see that the number of possible combinations of alleles is very large.
The column labeled “between markers” tells you the size class (“bin”) of the alleles, the column labeled “count” tells you the number of people in the sample population who had that allele. And, the column labeled “fraction” tells you the frequency of that bin in the sample.
INSTRUCTIONS: After you run your gel looking for your alleles at the D1S80 locus shade in the appropriate box to facilitate your probability calculation.
|
Size |
Control |
You**From Experiment
|
Suspect 1 |
Suspect 2 |
Evidence |
Size |
|
|
▀ |
|
|
|
|
|
▀ |
29 |
|
▀ |
|
|
|
|
|
▀ |
28 |
|
▀ |
|
|
|
|
|
▀ |
27 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
26 |
|
▀ |
|
|
|
|
|
▀ |
25 |
|
▀ |
|
|
|
|
|
▀ |
24 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
23 |
|
▀ |
|
|
|
|
|
▀ |
22 |
|
▀ |
|
|
|
|
|
▀ |
21 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
20 |
|
▀ |
|
|
|
|
|
▀ |
19 |
|
|
|
|
|
|
|
|
|
|
▀ |
▀ |
|
|
|
|
▀ |
18 |
|
|
|
|
|
|
|
|
|
|
▀ |
▀ |
|
|
|
|
▀ |
17 |
|
▀ |
|
|
|
|
|
▀ |
16 |
|
▀ |
|
|
▀ |
|
▀ |
▀ |
15 |
|
▀ |
|
|
|
|
|
▀ |
14 |
|
▀ |
|
|
|
▀ |
|
▀ |
13 |
|
▀ |
|
|
|
|
|
▀ |
12 |
|
|
|
|
|
▀ |
|
|
|
|
▀ |
|
|
|
|
|
▀ |
11 |
|
▀ |
|
|
|
|
|
▀ |
10 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
9 |
|
▀ |
|
|
▀ |
|
▀ |
▀ |
8 |
|
▀ |
|
|
|
|
|
▀ |
7 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
6 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
5 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
4 |
|
▀ |
|
|
|
|
|
▀ |
3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
2 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
1 |
D1S80 Locus
Before you begin your probability calculations take a careful look at all 5 autoradiographs provided to you.
Describe any trends you notice below.
Now you need to examine each autoradiograph and determine the frequency of the genotype for each allele for the 3 students present at the crime scene. Place your work in Table 1, Table 2, and Table 3.
Once you are determined the
probability of each genotype you are ready to determine the probability of each
person having the specific genetic make-up when multiple alleles are
considered. Record your work in table a, table b, and table c.
|
Size |
Control |
You |
Suspect 1 |
Suspect 2 |
Evidence |
Size |
|
|
▀ |
|
|
|
|
|
▀ |
29 |
|
▀ |
|
|
|
|
|
▀ |
28 |
|
▀ |
|
|
|
|
|
▀ |
27 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
26 |
|
▀ |
|
|
|
|
|
▀ |
25 |
|
▀ |
|
|
|
|
|
▀ |
24 |
|
|
|
|
▀ |
|
▀ |
|
|
|
▀ |
|
|
|
|
|
▀ |
23 |
|
▀ |
|
|
|
|
|
▀ |
22 |
|
▀ |
|
|
|
|
|
▀ |
21 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
20 |
|
▀ |
|
|
|
|
|
▀ |
19 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
18 |
|
|
|
|
▀ |
▀ |
▀ |
|
|
|
▀ |
|
|
|
|
|
▀ |
17 |
|
▀ |
|
|
|
|
|
▀ |
16 |
|
▀ |
|
|
|
|
|
▀ |
15 |
|
▀ |
|
|
|
|
|
▀ |
14 |
|
▀ |
|
|
|
|
|
▀ |
13 |
|
▀ |
|
|
|
|
|
▀ |
12 |
|
|
|
|
|
|
|
|
|
|
▀ |
▀ |
|
|
|
|
▀ |
11 |
|
▀ |
|
|
|
|
|
▀ |
10 |
|
|
|
▀ |
|
▀ |
|
|
|
|
▀ |
|
|
|
|
|
▀ |
9 |
|
▀ |
▀ |
|
|
|
|
▀ |
8 |
|
▀ |
|
|
|
|
|
▀ |
7 |
|
|
|
▀ |
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
6 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
5 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
4 |
|
▀ |
|
|
|
|
|
▀ |
3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
2 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
1 |
Examine the autoradiogram on the left. Identify the alleles present in each sample and the frequency at which each allele should appear in a population by using the data provided on the right. Record you information in the Data Table 1
|
Size |
Control |
You |
Suspect 1 |
Suspect 2 |
Evidence |
Size |
|
|
▀ |
|
|
|
|
|
▀ |
29 |
|
▀ |
|
|
|
|
|
▀ |
28 |
|
▀ |
|
|
|
|
|
▀ |
27 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
26 |
|
▀ |
|
|
|
|
|
▀ |
25 |
|
▀ |
|
|
|
|
|
▀ |
24 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
23 |
|
▀ |
|
|
|
|
|
▀ |
22 |
|
▀ |
|
|
|
|
|
▀ |
21 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
20 |
|
▀ |
|
|
|
|
|
▀ |
19 |
|
|
|
|
▀ |
|
▀ |
|
|
|
▀ |
|
|
|
|
|
▀ |
18 |
|
|
▀ |
|
|
|
|
|
|
|
▀ |
|
|
|
▀ |
|
▀ |
17 |
|
▀ |
|
|
|
|
|
▀ |
16 |
|
▀ |
|
|
|
▀ |
|
▀ |
15 |
|
▀ |
|
▀ |
|
|
|
▀ |
14 |
|
▀ |
|
|
▀ |
|
▀ |
▀ |
13 |
|
▀ |
|
|
|
|
|
▀ |
12 |
|
|
|
▀ |
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
11 |
|
▀ |
|
|
|
|
|
▀ |
10 |
|
|
▀ |
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
9 |
|
▀ |
|
|
|
|
|
▀ |
8 |
|
▀ |
|
|
|
|
|
▀ |
7 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
6 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
5 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
4 |
|
▀ |
|
|
|
|
|
▀ |
3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
2 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
1 |
Examine the autoradiogram on the left. Identify the alleles present in each sample and the frequency at which each allele should appear in a population by using the data provided on the right. Record you information in the Data Table 1

Examine the autoradiogram on the left. Identify the alleles present in each sample and the frequency at which each allele should appear in a population by using the data provided on the right. Record you information in the Data Table 1
|
Size |
Control |
You |
Suspect 1 |
Suspect 2 |
Evidence |
Size |
|
|
▀ |
|
|
|
|
|
▀ |
29 |
|
▀ |
▀ |
|
|
|
|
▀ |
28 |
|
▀ |
|
|
|
|
|
▀ |
27 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
26 |
|
▀ |
|
|
▀ |
|
▀ |
▀ |
25 |
|
▀ |
|
|
|
▀ |
|
▀ |
24 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
23 |
|
▀ |
|
|
|
|
|
▀ |
22 |
|
▀ |
|
|
|
|
|
▀ |
21 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
20 |
|
▀ |
|
|
|
|
|
▀ |
19 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
18 |
|
|
|
|
▀ |
|
▀ |
|
|
|
▀ |
|
|
|
▀ |
|
▀ |
17 |
|
▀ |
|
|
|
|
|
▀ |
16 |
|
▀ |
|
|
|
|
|
▀ |
15 |
|
▀ |
|
|
|
|
|
▀ |
14 |
|
▀ |
|
|
|
|
|
▀ |
13 |
|
▀ |
|
|
|
|
|
▀ |
12 |
|
|
|
▀ |
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
11 |
|
▀ |
|
|
|
|
|
▀ |
10 |
|
|
▀ |
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
9 |
|
▀ |
|
|
|
|
|
▀ |
8 |
|
▀ |
|
|
|
|
|
▀ |
7 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
6 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
5 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
4 |
|
▀ |
|
|
|
|
|
▀ |
3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
2 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
1 |

Examine the autoradiogram on the left. Identify the alleles present in each sample and the frequency at which each allele should appear in a population by using the data provided on the right. Record you information in the Data Table 1
|
Size |
Control |
You |
Suspect 1 |
Suspect 2 |
Evidence |
Size |
|
|
▀ |
|
|
|
|
|
▀ |
29 |
|
▀ |
|
▀ |
|
|
|
▀ |
28 |
|
▀ |
|
|
|
|
|
▀ |
27 |
|
|
▀ |
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
26 |
|
▀ |
|
▀ |
▀ |
|
▀ |
▀ |
25 |
|
▀ |
|
|
|
|
|
▀ |
24 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
23 |
|
▀ |
|
|
|
|
|
▀ |
22 |
|
▀ |
|
|
|
|
|
▀ |
21 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
20 |
|
▀ |
|
|
|
|
|
▀ |
19 |
|
|
▀ |
|
|
▀ |
|
|
|
|
▀ |
|
|
|
|
|
▀ |
18 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
▀ |
▀ |
▀ |
▀ |
17 |
|
▀ |
|
|
|
|
|
▀ |
16 |
|
▀ |
|
|
|
|
|
▀ |
15 |
|
▀ |
|
|
|
|
|
▀ |
14 |
|
▀ |
|
|
|
|
|
▀ |
13 |
|
▀ |
|
|
|
|
|
▀ |
12 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
11 |
|
▀ |
|
|
|
|
|
▀ |
10 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
9 |
|
▀ |
|
|
|
|
|
▀ |
8 |
|
▀ |
|
|
|
|
|
▀ |
7 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
6 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
5 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
4 |
|
▀ |
|
|
|
|
|
▀ |
3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
2 |
|
|
|
|
|
|
|
|
|
|
▀ |
|
|
|
|
|
▀ |
1 |
TABLE 1
|
Locus |
Allele |
Evidence matches suspect 1? |
Between marker # __ and marker # __ |
Frequency of Allele |
Frequency of Genotype= 2ab |
1 out of ____ people will display this genotype |
|
D2S44 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D1S7 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D10S26 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D4S139 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D1S80 |
1 |
|
|
|
|
|
|
2 |
|
|
|
According the rule of multiplication, the probability of two or more independent events occurring in combination is the product of the two or more independent event probabilities. Calculate the probability for the multi-locus genotype for two loci. Do this by multiplying together the frequencies of each of the two loci.
TABLE A
|
Locus |
Frequency of Genotype |
Multi-locus probability |
1 out of ____ people will display these loci in combination |
|
D2S44 |
|
N/A |
|
|
D2S44+D1S7 |
|
|
|
|
D2S44+D1S7+D10S26 |
|
|
|
|
D2S44+D1S7+D10S26+D4S139 |
|
|
|
|
D2S44+D1S7+D10S26+D4S139+D1S80 |
|
|
|
TABLE 2
|
Locus |
Allele |
Evidence matches you? |
Between marker # __ and marker # __ |
Frequency of Allele |
Frequency of Genotype= 2ab |
1 out of ____ people will display this genotype |
|
D2S44 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D1S7 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D10S26 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D4S139 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D1S80 |
1 |
|
|
|
|
|
|
2 |
|
|
|
According the rule of multiplication, the probability of two or more independent events occurring in combination is the product of the two or more independent event probabilities. Calculate the probability for the multi-locus genotype for two loci. Do this by multiplying together the frequencies of each of the two loci.
TABLE B
|
Locus |
Frequency of Genotype |
Multi-locus probability |
1 out of ____ people will display these loci in combination |
|
D2S44 |
|
N/A |
|
|
D2S44+D1S7 |
|
|
|
|
D2S44+D1S7+D10S26 |
|
|
|
|
D2S44+D1S7+D10S26+D4S139 |
|
|
|
|
D2S44+D1S7+D10S26+D4S139+D1S80 |
|
|
|
TABLE 3
|
Locus |
Allele |
Evidence matches you? |
Between marker # __ and marker # __ |
Frequency of Allele |
Frequency of Genotype= 2ab |
1 out of ____ people will display this genotype |
|
D2S44 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D1S7 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D10S26 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D4S139 |
1 |
|
|
|
|
|
|
2 |
|
|
|
|||
|
D1S80 |
1 |
|
|
|
|
|
|
2 |
|
|
|
According the rule of multiplication, the probability of two or more independent events occurring in combination is the product of the two or more independent event probabilities. Calculate the probability for the multi-locus genotype for two loci. Do this by multiplying together the frequencies of each of the two loci.
TABLE C
|
Locus |
Frequency of Genotype |
Multi-locus probability |
1 out of ____ people will display these loci in combination |
|
D2S44 |
|
N/A |
|
|
D2S44+D1S7 |
|
|
|
|
D2S44+D1S7+D10S26 |
|
|
|
|
D2S44+D1S7+D10S26+D4S139 |
|
|
|
|
D2S44+D1S7+D10S26+D4S139+D1S80 |
|
|
|
Results and Discussion
References